µManager is software for control of microscopes. It works with almost all microscopes, cameras and peripherals on the market, and provides an easy to use interface that lets you run your microscopy-based experiments. µManager runs as a plugin to ImageJ, is Open Source, and is free. µManager is developed in Ron Vale’s laboratory at UCSF and is funded by an NIH grant R01-EB007187 from the National Institute of Biomedical Imaging and Bioengineering (NBIB).
µManager serves a double purpose:
- It is a complete image acquisition and microscope control package, available for Windows, Mac and Linux, easy to install and configure right “out-of-the-box”, with built-in functionality and user interface for most common tasks performed in a Life Science laboratory.
- It also is a software framework for implementing advanced and novel imaging procedures, extending functionality, customization and rapid development of specialized imaging applications.
The main features of the µManager application are:
- Cross-platform code-base (Windows, Mac and Linux).
- Full integration with ImageJ, a widely used, freely available image processing package, developed at NIH.
- Intuitive user interface and simple installation procedure.
- Maximally extensible through support for scripting and dynamic programming languages.
- Maximum speed and efficiency in all control paths to allow for very high speed acquisition rates
- Hardware abstraction interface to allow writing hardware independent user interfaces and scripts for all devices used in the automated microscope, regardless of type, manufacturer and native drivers.
- Programmatic interfaces to 3rd-party analysis environments (such as Matlab) to enable “intelligent data collection” – i.e. analysis driven acquisition.
Software Architecture
The µManager software architecture is structured in three basic layers: Graphical User Interface (GUI), Core Services (MMCore) and Device Adapters. Each layer is largely independent of other layers and the architecture relies on two public interfaces, one on the GUI side and one on the device side, to bind all components into a single application. In addition to application layers the software also incorporates a scripting environment (editor and command interpreter) to extend functionality.
The foundation of the software is the “MMCore” module, written in C++, which controls and synchronizes various devices (cameras, shutters, stages, etc.) using plug-in “Adapter” modules. The MMCore interface exposes a generalized, abstract command set of the automated microscope that can be accessed from many different programming environments (C++, Java, Matlab, Python, Perl and others).
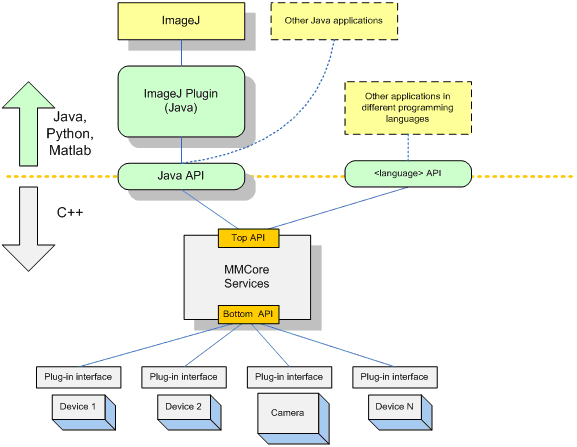
Figure 1, Block diagram of the software architecture
Besides core module MMCore, the µManager software consists of the following components:
- GUI front-end for interactive work with the integrated microscope, implemented in Java as an ImageJ plugin.
- Set of independent device adapter modules for particular hardware devices
- Scripting environment for writing automated procedures and executing command-line control commands
Core Services
The central component is the Core Services Module (MMCore) - a hardware abstraction layer providing services common to all automated microscopes. It also acts as insulation between the intricacies of particular device drivers and the user interface. MMCore provides two programming interfaces: a “Top” API (from GUI to MMCore) and a “Bottom” API (from device drivers to MMCore). The “Top API” gives the designer of the GUI the ability to relatively easily implement user interfaces and entire applications that are essentially hardware independent. The “Bottom API” is a C/C++ plugin interface allowing the programmer to support particular devices with a minimal amount of coding effort and minimal understanding of how the entire system works.
MMCore provides the following device-independent services:
- Flexible hardware configuration facilities to support defining, naming and adding new devices
- Run-time device discovery and on-the-fly loading and unloading of devices
- Automatic metadata generation and image annotation including the entire state of the hardware system at the time of acquisition
- Device synchronization (e.g. camera acquisition with stages and filter wheels)
- High resolution timing (for high speed sequence acquisition)
- Saving image files with automatic name generation and acquisition directory management
- Commonly used acquisition modes such as multiple channel, Z-stack and time-lapse
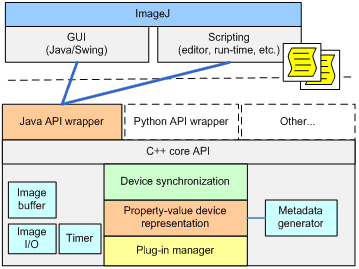
Figure 2, MMCore module block diagram
General design features
The most important design guidelines in our project were modularity and extensibility. By following these broad principles we tried to circumvent serious problems one encounters with the currently available commercial software: monolithic structure and lack of flexibility. At the same time we wanted to create a microscopy imaging application that will be easy to install and a pleasure to use for a broad range of users, especially non-programmers. Ease-of-use and flexibility are often perceived as conflicting requirements, but we think that these two goals can be achieved simultaneously without compromise. The following sections briefly touch upon the design principles and tools we used in creating the µManager software.
Separation of User Interface and Services
The µManager software and derived applications are designed around the client-server paradigm. In other words, the front-end GUI is strictly separated from the hardware control functionality by an abstract automated microscope API. All commands follow a simple request-response model, with the GUI sending requests and the MMCore servicing them.
Client-server design enables easy and robust implementation of a distributed software system where multiple clients and servers communicate with each other over the network. Client-Server design also decouples the GUI from the functional module, making development and extending much easier than with monolithic design. The current version of µManager does not support network or multi-user access, but the client/server design makes such extensions possible without significant changes to the existing software.
Portability
We recognize the fact that complete platform neutrality is extremely difficult to achieve in practice and we focused our efforts on supporting three operating systems commonly used in Life Science laboratories: Windows, Linux and Mac OS X. The µManager code base is a true cross-platform system. The back-end is written in C++ and compiles with GNU tools on Mac/Linux and Microsoft Developer Studio (MSDEV) on Windows. For system-level functionality (threads, logging, timers, etc.) we rely on the boost C++ libraries. The front-end GUI is written in Java with SWING based user interface components.
Cross platform Graphical User Interface
Cross platform development is most challenging when it comes to building user interfaces. The Java GUI front-end combined with a C++ back-end has proven to be a reliable and agile solution for the µManager project. By being able to rapidly prototype and deploy user interfaces in the Eclipse IDE, equipped with Windows Designer, we increased our ability to deliver new features quickly.
Also, taking advantage of extensive ImageJ libraries and 3rd party plugins turned out to be crucial for the success of the project. µManager can run either as a standalone application or as an ImageJ plugin. Tight integration with ImageJ enables us to switch to the ImageJ user interface whenever we need functionality that is not available in µManager.
Interfaces for Multiple Dynamic Languages
The core µManager API is designed to be used from dynamic languages such as Python and Ruby as well as from C++ and Java. The bindings for each programming language are generated automatically from the C++ header files, using the SWIG interface generator. Practically any programming language supported by SWIG can be used for programming µManager. Including support for each additional programming language in the standard software build requires a relatively small effort. µManager operation outside of its standard GUI and ImageJ environment was successfully demonstrated by controlling a fully automated microscope and digital camera from Matlab, with a relatively simple set of commands.
Installation and Configuration
Installation of the software consists of a single installer package for Windows, while on Mac and Linux it involves only copying a folder containing the software onto the end-user’s computer. The system configuration depends on the actual hardware setup – attached devices must be specified in a configuration text file. Examples for various commonly used configurations are provided as well as documentation for editing configuration files. Once hardware devices are specified and initialized, the command set can be easily customized using device management features in the User Interface. For example, the user can configure simultaneous actions on multiple devices to be executed following a single command.
